Research
Research
Research in our group focuses on understanding how microorganisms utilize sophisticated purpose-built nanomachines for transferring genetic material and effector molecules across membranes, and facilitate adherence to a variety of surfaces. Understanding of how these nanosystems are assembled from their component proteins, as well as their effects on infection and the development of resistance strategies, is critical to the development of more streamlined approaches to dealing with infection and drug resistance in pathogenic organisms. In addition, understanding how these systems function at a structural level is central to the development of biological systems for applications as biosensors.
Our primary research tool is high resolution X-ray crystallography, however we are interested in the correlation of multiple biophysical techniques in the examination of protein structure and function. This includes the use of Nuclear Magnetic Resonance (NMR), Small Angle X-ray Scattering (SAXS), Multi-Angle Light Scattering (MALS), Electron Microscopy (EM), X-ray fibre diffraction, and Hydrogen Deuterium Exchange Mass Spectrometry (HDX-MS), which can all provide significant insights into protein structure and dynamics and can complement well designed functional studies. Each distinct methodology has its own respective strengths, and the coupling of these methods can only benefit our understanding into the relation between protein structure and biochemical function.
As we are a crystallography lab, many of our computers are Linux-based. There are a variety of reasons for this, including security, data handling, crystallographic software developments etc. Follow this link to get a small primer on Linux, but of course you can search the internet as well…there are numerous sites out there talking about Linux.
Structural and Functional Studies of Proteins Involved in Bacterial Conjugation
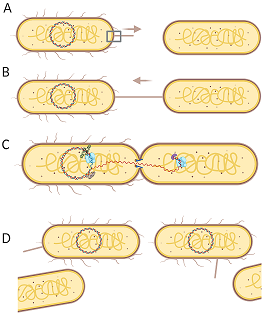
Bacterial conjugation is the process by which DNA can be transferred from one bacterium (donor) to another (recipient). This process enables the distribution of genetic material throughout a bacterial population, and is a mechanism for bacterial evolution and adaptation to unique environments. It is also a mechanisms by which genes encoding antibiotic resistance and other virulence factors are spread through a bacterial population. We are interested in the structures, dynamics and interactions of proteins involved in assembly the type IV secretion systems utilized in bacterial conjugation; our current focus is on the F plasmid from E. coli, as well as other important bacterial conjugative systems.
Pilin-Derived Protein Nanotubes
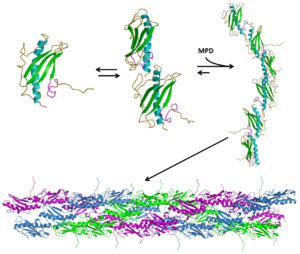
We are currently focusing on understanding the mechanisms of how pilin-dervied protein nanotubes (PNTs) oligomerize from the monomeric protein precursor, using the type IV pilins of Pseudomonas aeruginosa as a model, and looking to adapt them for applications in bionanotechnology. We are also interested in otehr pilin/PNT systems, including those from Coxiella burnetii, Francisella tularensis and Burkholderia pseudomalei to understand the similarities and differences between different pilins.